Hi, this is the website of the NanoBioPhotonix Lab in Tel-Aviv university.
The main focus of the group is Single-molecule genomics but we have activity also in development of new optical detection schemes and novel imaging techniques. We explore genomes utilizing tools and reagents from the realm of nano-technology. We try learning new things about these systems by zooming in on individuals - single cells, single chromosomes and single molecules. Research in the lab is highly multi and inter disciplinary and our team is composed of chemists, biologists and physicists who are interested in learning from each other and doing some great (but sometimes risky...) stuff at the very forefront of science.




News

Biosoft - Tsuba Meeting
17 Feb 2026
Dr. Shira Wisthal-Algor, Supervisor: Prof. Yuval Ebenstein
"Carboxymethylation Labeling of Unmethylated Epigenetics Sites"

Physical Chemistry Graduate Seminar Day
15 Feb 2026
Sigal Avraham, Phd Supervisor: Prof. Yuval Ebenstein
"Zooming in on the Epigenetics of Hematological Malignancies"

ICS89 - The 89 annual metting of the Israeli chemical society at TAU
10-11 Feb 2026
Organized by Prof. Yuval Ebenstein & Prof. Roee Amir
Sepcial Flash talk speaker from our lab - Shani Dvir:
"Mapping Specific Locations on DNA through Adenine Synthetic Modifications"

Hanxmas 2025 group meeting
28 Dec 2025
Wishing all the best year ever!

The 20th Biennial Cancer Biology Research Center Meeting 2025, Galile
17-19 Sep 2025
Prof. Ebenstein & the team presenting posters:
"Enzymatic profiling of cell-free DNA methylation for detection and monitoring of lung cancer"
"Minimaly invaise Lymphoma diagnostics using epigenetic biomarkers"
"MiRACLE: Single-molecule microRNA detection for early disease diagnostics of diffuse large B-cell Lymphoma"
"Non-invasive DNA methylation assay distinguishes between monoclonal gammopathy of Undetermined Signficance (MGUS), Smoldering Myeloma (SMM) and Multiple Myeloma (MN)"
"Adenine modification for high throughput Multi-Omic mapping of DNA"
"Dual-color microarray profiling of 5mc and 5hmc reveals epigentic signtures in Myeloproliferative Neoplasmas (MPN)"

LMI Retreat 2025, Nazareth
7-8 Sep 2025

Congratulations!
12 Jul 2025
Prof. Yuval Ebenstein is among ten senior Israeli researchers who have been awarded the prestigious ERC Proof of Concept (PoC) grant by the European Research Council. These grants, announced Monday, support the commercialization of academic research and are given only to previous ERC grant recipients.

Woodstock.Bio2 Meeting in Prague, CZ
11 Jun 2025

Passover Picnic - Botanical Garden
7 Apr 2025

2 New publications in NAR Genomics & Bioinformatic
12 March 2025
Long-read structural and epigenetic profiling of a kidney tumor-matched sample with nanopore sequencing and optical genome mapping
&
Single-molecule toxicogenomics: Optical genome mapping of DNA-damage in nanochannel arrays

MIXiii Health-Tech.IL 2025 - IT'S SHOW TIME !
5 Mar 2025

Welcome to the new Nano building!
9 Dec 2024
The Jan Koum Center for Nanoscience and Nanotechnology is a research facility focused on cutting-edge research and technological advancement.
.png)
It's all about Chemistry...
The podcast 'Tel Aviv-360' hosting Prof. Yuval Ebenstein
17 Nov 2024
Listen to this speacial episode with Prof. Ebenstein - What is 'Epigenetics' and How Do Our Emotions Affect DNA?

Group meeting in the outdoors
11 Nov 2024
Read more

2 New publications in Blood jurnal
5 Nov 2024
Targeted, Low Cost Epigenetic Microarray for AML and MDS Detection & Detection of B Cell Lymphomas By a Simple Epigenetic Blood Test
Single-Molecule Sensors and NanoSystems International
Conference- S3IC Paris 2024
28-30 Oct 2024
The presentation of the paper- Single-molecule Toxicogenomics: Optical Genome Mapping of DNA-damage in nanochannel arrays by Prof. Ebenstein & the presentation of the paper-DeepQR: Single-molecule QR codes provide extreme multiplexing for optical gene-expression analysis by Dr. Jonathan Jeffet
LMI Students Seminar: Lanna & Jonathan Presenting
18 July 2024
Lanna Bery presenting 'High resolution spectral microscopy'
Jonathan Jeffet presenting 'DeepQR: single-molecule QR codes for optical gene-expression analysis'
Swedish Microfluidics in Life Sciences Conference – 4-5 June 2024
4 June 2024
The 2024 SMILS conference will be jointly hosted by the Department of Life Sciences at Chalmers and the Department of Physics at the University of Gothenburg.
One of the plenary speakers is Prof. Yuval Ebenstein.
Abstract published in Medrxiv
3 May 2024
Oxford Nanopore Technology (ONT) based methylation sequencing is increasingly recognized for its rapid and accurate classification of brain tumors. A process that is crucial for optimal patient treatment. However, widespread clinical utility is currently limited by the need for fresh-frozen biopsies and not the standard-of-care formalin-fixed, paraffin-embedded (FFPE) samples. Our study explores the impact of FFPE on DNA methylation and presents a developed and validated protocol for ONT-based FFPE tumor classification. We present a practical solution for precise brain tumor diagnoses in routine clinical settings and facilitating timely treatment decisions at the point of care and without interfering with operating room standards.
Abstract published in Biorxiv
1 April 2024
Carcinogenesis often involves significant alterations in the cancer genome architecture, marked by large structural and copy number variations (SVs and CNVs) that are difficult to capture with short-read sequencing. Traditionally, cytogenetic techniques are applied to detect such aberrations, but they are limited in resolution and do not cover features smaller than several hundred kilobases. Optical genome mapping and nanopore sequencing are attractive technologies that bridge this resolution gap and offer enhanced performance for cytogenetic applications. These methods profile native, individual DNA molecules, thus capturing epigenetic information. We applied both techniques to characterize a clear cell renal cell carcinoma (ccRCC) tumor's structural and copy number landscape, highlighting the relative strengths of each method in the context of variant size and average read length. Additionally, we assessed their utility for methylome and hydroxymethylome profiling, emphasizing differences in epigenetic analysis applicability.
Congratulations! The proposal for an ERC Proof of Concept Grant:
MiRACLE, has been selected for funding
18 January 2024
We got 150,000 Euros for our project: Multiplexed microRNA detection platform for early diagnosis and patient management.
The 65th ASH Annual Meeting Abstracts - Abstract published in Blood
28 November 2023
Acute Myeloid Leukemia (AML) is marked by uncontrolled growth of undifferentiated myeloid cells. DNA methylation patterns in AML have been thoroughly researched, however, studies on hydroxy methylation patterns, indicating active DNA methylation erasure, are still evolving. Our principal goal was to detect differential methylation and hydroxy methylation regions between AML and non-malignant samples. We proposed that simultaneous profiling of blood cells and circulating cell-free DNA would offer more insights into the disease mechanism. Therefore, we combined these data sources to provide a comprehensive epigenetic picture, assuming that this dual approach could reveal new AML biomarkers and enhance our understanding regarding AML pathobiology.
Single-Molecule Sensors and NanoSystems International Conference – S3IC 2023, Barcelona
23 November 2023
The presentation of the abstract- "Machine learning based Singlemolecule Quantification of Circulating Micro RNA Mixtures"

Article published in ChemBioChem
17 October 2023
5-Methylcytosine and 5-hydroxymethylcytosine are epigenetic modifications involved in gene regulation and cancer. We present a new, simple, and high-throughput platform for multi-color epigenetic analysis. The novelty of our approach is the ability to multiplex methylation and de-methylation signals in the same assay. We utilize an engineered methyltransferase enzyme that recognizes and labels all unmodified CpG sites with a fluorescent cofactor. In combination with the already established labeling of the de-methylation mark 5-hydroxymethylcytosine via enzymatic glycosylation, we obtained a robust platform for simultaneous epigenetic analysis of these marks. We assessed the global epigenetic levels in multiple samples of colorectal cancer and observed a 3.5-fold reduction in 5hmC levels but no change in DNA methylation levels between sick and healthy individuals. We also measured epigenetic modifications in chronic lymphocytic leukemia and observed a decrease in both modification levels (5-hydroxymethylcytosine: whole blood 30 %; peripheral blood mononuclear cells (PBMCs) 40 %. 5-methylcytosine: whole blood 53 %; PBMCs 48 %). Our findings propose using a simple blood test as a viable method for analysis, simplifying sample handling in diagnostics. Importantly, our results highlight the assay‘s potential for epigenetic evaluation of clinical samples, benefiting research and patient management.

Article published in ACS sensors
4 October 2023
MicroRNAs (miRs) are small noncoding RNAs that regulate gene expression and are emerging as powerful indicators of diseases. MiRs are secreted in blood plasma and thus may report on systemic aberrations at an early stage via liquid biopsy analysis. We present a method for multiplexed single-molecule detection and quantification of a selected panel of miRs. The proposed assay does not depend on sequencing, requires less than 1 mL of blood, and provides fast results by direct analysis of native, unamplified miRs. This is enabled by a novel combination of compact spectral imaging and a machine learning-based detection scheme that allows simultaneous multiplexed classification of multiple miR targets per sample. The proposed end-to-end pipeline is extremely time efficient and cost-effective. We benchmark our method with synthetic mixtures of three target miRs, showcasing the ability to quantify and distinguish subtle ratio changes between miR targets.
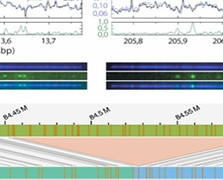
Article published in DNA Repair
5 July 2023
The human genome is continually exposed to various stressors, which can result in DNA damage, mutations, and diseases. Among the different types of DNA damage, single-strand lesions are commonly induced by external stressors and metabolic processes. Accurate detection and quantification of DNA damage are crucial for understanding repair mechanisms, assessing environmental impacts, and evaluating response to therapy. However, traditional techniques have limitations in sensitivity and the ability to detect multiple types of damage. In recent years, single-molecule fluorescence approaches have emerged as powerful tools for precisely localizing and quantifying DNA damage. Repair Assisted Damage Detection (RADD) is a single-molecule technique that employs specific repair enzymes to excise damaged bases and incorporates fluorescently labeled nucleotides to visualize the damage. This technique provides valuable insights into repair efficiency and sequence-specific damage. In this review, we discuss the principles and applications of RADD assays, highlighting their potential for enhancing our understanding of DNA damage and repair processes.

Article published in ACS nano
8 May 2023
Proteins and enzymes in the cell nucleus require physical access to their DNA target sites in order to perform genomic tasks such as gene activation and transcription. Hence, chromatin accessibility is a central regulator of gene expression, and its genomic profile holds essential information on the cell type and state. We utilized the E. coli Dam methyltransferase in combination with a fluorescent cofactor analogue to generate fluorescent tags in accessible DNA regions within the cell nucleus. The accessible portions of the genome are then detected by single-molecule optical genome mapping in nanochannel arrays. This method allowed us to characterize long-range structural variations and their associated chromatin structure. We show the ability to create whole-genome, allele-specific chromatin accessibility maps composed of long DNA molecules extended in silicon nanochannels.

Article published in Bioinformatics
1 March 2023
Efficient tapping into genomic information from a single microscopic image of an intact DNA molecule is an outstanding challenge and its solution will open new frontiers in molecular diagnostics. Here, a new computational method for optical genome mapping utilizing deep learning is presented, termed DeepOM. Utilization of a convolutional neural network, trained on simulated images of labeled DNA molecules, improves the success rate in the alignment of DNA images to genomic references.
The method is evaluated on acquired images of human DNA molecules stretched in nano-channels. The accuracy of the method is benchmarked against state-of-the-art commercial software Bionano Solve. The results show a significant advantage in alignment success rate for molecules shorter than 50 kb. DeepOM improves the yield, sensitivity, and throughput of optical genome mapping experiments in applications of human genomics and microbiology.

Article published in Nucleic acids research
9 September 2022
DNA methylation, specifically, methylation of cytosine (C) nucleotides at the 5-carbon position (5-mC), is the most studied and significant epigenetic modification. Here we developed a chemoenzymatic procedure to fluorescently label non-methylated cytosines in CpG context, allowing epigenetic profiling of single DNA molecules spanning hundreds of thousands of base pairs. We used a CpG methyltransferase with a synthetic S-adenosyl-L-methionine cofactor analog to transfer an azide to cytosines instead of the natural methyl group. A fluorophore was then clicked onto the DNA, reporting on the amount and position of non-methylated CpGs. We found that labeling efficiency was increased up to 2-fold by the addition of a nucleosidase, presumably by degrading the inactive by-product of the cofactor after labeling, preventing its inhibitory effect. We used the method to determine the decline in global DNA methylation in a chronic lymphocytic leukemia patient and then performed whole-genome methylation mapping of the model plant Arabidopsis thaliana. Our genome maps show high concordance with published bisulfite sequencing methylation maps. Although mapping resolution is limited by optical detection to 500–1000 bp, the labeled DNA molecules produced by this approach are hundreds of thousands of base pairs long, allowing access to long repetitive and structurally variable genomic regions.

Article published in Biophysical Reports
8 December 2021
Mapping DNA damage and its repair has immense potential in understanding environmental exposures, their genotoxicity, and their impact on human health. Monitoring changes in genomic stability also aids in the diagnosis of numerous DNA-related diseases, such as cancer, and assists in monitoring their progression and prognosis. Developments in recent years have enabled unprecedented sensitivity in quantifying the global DNA damage dose in cells via fluorescence-based analysis down to the single-molecule level. However, genome-wide maps of DNA damage distribution are challenging to produce. Here, we describe the localization of DNA damage and repair loci by repair-assisted damage detection sequencing (RADD-seq).

Article published in Bioinformatics
12 July 2021
While promoter methylation is associated with reinforcing fundamental tissue identities, the methylation status of distant enhancers was shown by genome-wide association studies to be a powerful determinant of cell-state and cancer. With recent availability of long reads that report on the methylation status of enhancer–promoter pairs on the same molecule, we hypothesized that probing these pairs on the single-molecule level may serve the basis for detection of rare cancerous transformations in a given cell population. We explore various analysis approaches for deconvolving cell-type mixtures based on their genome-wide enhancer–promoter methylation profiles.

Sapir won the Women of chemistry Forum Award
20 July 2021
Sapir won for her work on Long reads capture simultaneous enhancer–promoter methylation status for cell-type deconvolution
Congratulation

Sapir won the David and Paulina Trotsky Foundation Award
6 April 2021
Sapir won for her work on Single-cell level epigenetic analysis in distal regulatory elements
Congratulation

Noa won the Avital and Eran Rabani award
29 April 2021
Noa won this award for her work on Rapid-RADD DNA Damage Detection, Designed to be used as a method of quantifying DNA damage for multiple DNA samples in one.
Congratulation

Goodbye from Tamar
29 April 2021
Saying good bye to Dr. Tamar Shahal who will be moving on in her Career.
Good Luck!

Jonathan won the excellence award for teaching and research
3 May 2021
Jonathan has won the Excellence award for teaching and research from the school of Physics and Astronomy
congratulations

Article published in Essays Biochem
16 April2021
The human genome contains multiple layers of information that extend beyond the genetic sequence. In fact, identical genetics do not necessarily yield identical phenotypes as evident for the case of two different cell types in the human body. The great variation in structure and function displayed by cells with identical genetic background is attributed to additional genomic information conten

Article published in BiorXiv
29 January 2021
Motivation While promoter methylation is associated with reinforcing fundamental tissue identities, the methylation status of distant enhancers was shown by genome-wide association studies to be a powerful determinant of cell-state and cancer. With recent availability of long-reads that report on the methylation status of enhancer-promoter pairs on the same molecule, we hypothesized that probing these pairs on the single-molecule level may serve the basis for detection of rare cancerous transformations in a given cell population. We explore various analysis approaches for deconvolving cell-type mixtures based on their genome-wide enhancer-promoter methylation profiles.

Sigal presented in the CBRC 3rd virtual seminar
14 October 2020
Rapid quantification of 5mC and 5hmC on multi-sample array slides

Gal presented a poster in the Retreat of the Edmond J. Safra Center for Bioinformatics
24 May 2020
Using nanopore sequencing to detect base modifications

Jonathan presented in SPAOM2020
24 November 2020
Multi-Model Single-Molecule Imaging with continuously controlled spectral-resolution Microscopy

Article published in Nanoscale
1 October 2020
Non-DNA labels are key components for the construction of functional DNA nanostructures. Here, we present a method to graft covalent labels onto DNA origami nanostructures in an enzymatic one-pot reaction. The DNA methyltransferase M.TaqI labels the DNA nanostructures with azide groups, which serve as universal attachment points via click chemistry. Direct labeling with fluorescent dyes is also demonstrated. The procedure yields structures with high fluorescence intensities and narrow intensity distributions. In combination with UV crosslinking it enables the creation of temperature-stable, intense fluorescent beacons.

Article published in Translational Oncology
8 July 2020
Ionizing radiation (IR) is a common mode of cancer therapy, where DNA damage is the major reason of cell death. Here, we use an assay based on fluorescence imaging of single damaged DNA molecules isolated from radiated lymphocytes, to quantify IR induced DNA damage. The assay uses a cocktail of DNA-repair enzymes that recognizes and excises DNA lesions and then a polymerase and a ligase incorporate fluorescent nucleotides at the damage sites, resulting in a fluorescent “spot” at each site. The individual fluorescent spots can then be counted along single stretched DNA molecules and the global level of DNA damage can be quantified. Our results demonstrate that inclusion of the human apurinic/apyrimidinic endonuclease 1 (APE1) in the enzyme cocktail increases the sensitivity of the assay for detection of IR induced damage significantly. This optimized assay also allowed detection of a cooperative increase in DNA damage when IR was combined with mild hyperthermia, which is sometimes used as an adjuvant in IR therapy. Finally, we discuss how the method may be used to identify patients that are sensitive to IR and other types of DNA damaging agents.
24 June 2020
Knowing the amount and type of DNA damage is of great significance for a broad range of clinical and research applications. However, existing methods either lack in their ability to distinguish between types of DNA damage, or are limited in their sensitivity and reproducibility. The method described herein enables rapid and robust quantification of type-specific single-strand DNA damage. The method is based on Repair-Assisted Damage Detection (RADD) by which fluorescent nucleotides are incorporated into DNA damage sites using type-specific repair enzymes. Up to 90 DNA samples are then deposited on a multi-well glass slide, and analyzed by a conventional slide scanner for quantification of DNA damage levels. Accurate and sensitive measurements of oxidative or UV-induced DNA damage levels and repair kinetics are presented for both in-vitro and in-vivo models.Add News Story here
2 September 2019
Herein we present an assay allowing concurrent detection of oxidative DNA damage and photoproducts. We apply DNA repair enzymes specific for each lesion type to incorporate spectrally distinct fluorescent nucleotides, enabling simultaneous quantification of the lesions on individual DNA molecules. We follow the repair of both damage types in skin cells exposed to artificial sunlight.

3 May 2019
Check it out! we were mentioned on NATURE
Article Published in Analytica Chimica Acta: "Hypersensitive quantification of global 5-hydroxymethylcytosine by chemoenzymatic tagging"
22 August 2018
One of the challenges associated with detecting 5hmC levels is its extremely low content, especially in blood. Detecting 5hmC levels in blood samples for diagnosis of leukemia and other blood malignancies presents a unique challenge. To overcome these difficulties we introduce a simple chemoenzymatic method for specifically tagging 5hmC, resulting in an eight-fold increase in detection sensitivity. We demonstrate that we could quantitatively detect 5hmC levels in various human tissues, including blood samples from healthy individuals and leukemia patients, using the most basic quadrupole mass-analyzer instrument and only 200 ng of DNA. The limit of detection (LOD) of our technique is 0.001% 5hmC from 300 ng DNA, sufficient for future mass-spectroscopy based diagnostics of diseases associated with low 5hmC levels such as leukemia
Article Published in "ACS Nano": Epigenetic Optical Mapping of 5-Hydroxymethylcytosine in Nanochannel Arrays
20 June 2018
The epigenetic mark 5-hydroxymethylcytosine (5-hmC) is linked to gene regulation, development, and disease. In particular, its levels dramatically decline in many cancers, potentially serving as an epigenetic biomarker.
We present a long-read, highly sensitive single-molecule mapping technology that generates hybrid genetic/epigenetic profiles of native chromosomal DNA. The single-molecule concept provides information on the distribution and coexistence of 5-hmC signals at multiple genomic loci on the same genomic DNA molecule, revealing long-range correlations and cell-to-cell epigenetic variation.
To view the paper in open access click the "Read More" button below
Article Published in "Nucleic Acids Research": Selective nanopore sequencing of human BRCA1 by Cas9-assisted targeting of chromosome segments (CATCH)
21 May 2018
Here, we optimized Cas9-Assisted Targeting of CHromosome segments (CATCH) for nanopore sequencing of the breast cancer gene BRCA1. The CATCH enrichment scheme only requires knowledge of the target flanking sequence for Cas9 cleavage while providing contiguous data across both coding and non-coding sequence and holds promise for characterization of complex disease-related or highly variable genomic regions.
To view the paper in open access click the "Read More" button below
Article Published in "DNA Repair": Broad spectrum detection of DNA damage by Repair Assisted Damage Detection (RADD)
27 April 2018
Numerous methods to characterize the formation of DNA adducts and their retention for risk assessment have been developed. Here, we describe a new methodology, Repair Assisted Damage Detection (RADD), which utilizes a DNA damage processing repair enzyme cocktail to detect and modify sites of DNA damage for a subsequent gap filling reaction that labels the DNA damage sites.
To view the paper click the "Read More" button below

Congratulations to Dr. Tamar Shahal for Best Poster Award in Isranalytica 2018!
January 24, 2018

Noa's participation in a conference of the XIN project at Tsinghua University, China
September 2017
Noa presented an abstract in a conference of the XIN project at Tsinghua University, Beijing, China. Noa used single-molecule fluorescence imaging to detect chemical changes in individual DNA molecules and showed she can detect colon and blood cancer with extreme sensitivity.
Her work was recently published in the journal Clinical Epigenetics: "Single-molecule quantification of 5-hydroxymethylcytosine for diagnosis of blood and colon cancers".
Click the "Read More" button to see the paper.
Dima's working visit to the University of Minnesota
August 2017
Dima has recently received the Prof. Rahamimoff Travel Grant for Young Scientists from the BSF United States – Israel Binational Foundation, for a working visit to University of Minnesota at Prof. Kevin Dorfman's lab. During his stay from 26.7.17-24.8.17, he was trained on fabricating nano-fluidic devices in fused silica that utilizing electricity enable the manipulation and imaging of single DNA molecules.
In the figure: DNA molecules labeled with YOYO-1 as imaged in a fabricated device. Each white line represents an individual DNA molecule in a 100nm channel.

Gil's participation in EUROCARB 2017, Barcelona
June 2017
The EUROCARB 2017 program consolidated the interplay between the chemistry and biology communities, and reinforced the needed interaction between glycochemistry, glycobiology, and applied glycosciences.
Gil gave a talk titled "UDP-sugar derivative as a key substrate for DNA epigenetic modification labeling"
Article published in "Clinical Epigenetics": Single-molecule quantification of 5-hydroxymethylcytosine for diagnosis of blood and colon cancers
July 14, 2017
Using a single-molecule approach, we observed a significantly reduced level of 5hmC in blood and colon cancers, and could distinguish between colon tumor and colon tissue adjacent to the tumor based on the global levels of this molecular biomarker.
To view the paper in open access click the "Read More" button below

Participation in Xin Innovation Forum 2017
July 4, 2017
Congrats to Noa for being invited to participate in the Xin Innovation Forum 2017, which will be held in Beijing this September!

Dima is awarded the Prof. Rahamimoff Travel Grant for Young Scientists of the US-Israel Binational Science Foundation (BSF)
July 4, 2017
Congratulations to our very own Dima, for being selected to receive the Prof. Rahamimoff Travel Grant for Young Scientists of the US-Israel Binational Science Foundation (BSF)
Check out our attention score for the CATCH paper in biorxivs!
March 14, 2017
Our innovative targeting method - CHromosome segments Assisted targeting of Cas9 (CATCH) for long read nanopore sequencing and optical mapping - received high attention score in bioRxiv!
- It's in the top 5% of all research outputs scored by Altmetric
- Has high attention score compared to outputs of the same age and source (98th percentile)
- ans high attention score compared to outputs of the same age (97th percentile)

Collaboration with the Jaroslav Dolezel lab from the Institute of Experimental Botany
November 14, 2016
We have recently welcomed Zuzana from the Centre of Plant Structural and Functional Genomics in the Czech Republic, to work on the CATCH technique in our lab.

Comgrats to Dima and Jonthan for winning 3rd and 1st poster prizes at the i-core meeting!
September 27, 2016

Congratulations to Tslil for winning Best Poster Award in Nano-Israel 2016!
February 23, 2016

Sizing femtogram amounts of dsDNA by single-molecule counting
September 13, 2015
Modern molecular-biology applications raise renewed interest in sizing minute-amounts of DNA. In this work we utilize single-molecule imaging with in situ size calibration to accurately analyze the size and mass distribution of DNA samples. We exploit the correlation between DNA length and its fluorescence intensity after staining in order to assess the length of individual DNA fragments by fluorescence microscopy. Synthetic reference DNA standards are added to the investigated sample before staining and serve as internal size calibrators, supporting a robust assay for accurate DNA sizing. Our results demonstrate the ability to reconstruct the exact length distribution in a complex DNA sample by sizing a subset containing only femtogram amounts of DNA, thus, outperforming microfluidic gel electrophoresis which is the currently accepted gold standard. This assay may find useful applications for genetic analysis where the exact size distribution of DNA molecules is critical and the availability of genetic material is limited.

Baby Boom! Congratulations to the Fishmans & the Shahals!
September 12, 2015
Congratulations to Tamar Shahal & Sivan Fishman,
on the safe arrival of your baby boy & baby girl (respectively) into the world, and in a 5-minute difference!
May Babyhood be filled with lots of joy and make lots of wonderful memories.
We wish you all the best!

CRISPR-Cas9- Assisted Targeting of CHromosome segments (CATCH)
July 21 2015
Cas9 can be engineered to cut specific genomic loci. In our new paper accepted to Nature Communications, two such enzymes are used in-vitro in order to cut-out any genomic region of interest for further downstream applications with the selected region.
Together with our colleagues from Prof. Ting Zhu’s lab in Tsinghua University of Beijing we utilize this approach to isolate specific gene clusters up to 200 kbp and clone them into BAC vectors.

Major research bottleneck solved!
July 3rd 2015
In a new paper in ChemBioChem we report on a One-Pot Chemoenzymatic Cascade for Labelling of the Epigenetic Marker 5-Hydroxymethylcytosine. In a joint project with Dr. Micha Fridman from the organic chemistry department we found an elegant way of utilizing enzymes to assist in the preparation of a key reagent for 5-hmC labeling.

In a new paper we show how DNA mapping can distinguish between viruses
May 15, 2015
Research of microbiological environments displays a growing scientific interest, unmasking their great variability and specifically characterizing their population has applications for public health and biotechnological development. In our recent paper "Bacteriophage strain typing by rapid single molecule analysis" published in the journal Nucleic Acid Research, we show that we can identify single short genomes of bacteriophages. We fluorescently label the genomes in a sequence specific manner and measure the amplitude modulations of the fluorescent signal along stretched DNA. These modulations displays organism specific pattern and can refer as finger print for identification. This research can be applied to study and characterization of biological samples with unknown contents.

Our lab was awarded 1.1 million Euros for developing next generation diagnostic tools
April 15, 2015
We are the coordinators of the BeyonSeq consortium!
With only 1.9% success rate, we were lucky to be awarded six and a half million Euros to be shared between 7 research groups, to develop single-molecule diagnostic technologies in various areas.
The unifying theme is the analysis of individual DNA molecules.

Collaboration published in Nature Nanotechnology
March 16, 2015
A paper titled "Light-emitting self-assembled peptide nucleic acids exhibit both stacking interactions and Watson–Crick base pairing" was published by The Gazit lab and we had the pleasure to help. While imaging these interesting self assembled structures we found that they emit light across the visible spectrum. We also helped analyzing the growth dynamics and with the qualitative model for the intersting light emission effects

High throughput quantification of 5hmC - Published in Ananlytical Chemistry
August 5, 2014
We further developed our method for optical detection of 5hmC to be compatible with measurements on a multiwell plate. Now ~350 samples can be measured simultaneously. We demonstrate a measurement of 190 mouse tissue samples.

Lighting up individual DNA damage sites by in-vitro repair synthesis
May 13, 2014
Our work on direct visualization of DNA damage was just accepted to JACS.
We repair extracted genomic DNA in a tube containing a cocktail of repair enzymes and fluorescent nucleotides. Damaged DNA lights up as it is repaired and is visualized as fluorescent spots along the DNA molecules.

Toward Single-Molecule Optical Mapping of the Epigenome
Our new review is out in ACS NANO. We show exciting preliminary results of engineered 5hmC patterns on Lambda DNA. Individual molecules are stretched and imaged in silicon nano-channel arrays.

I've been told that it's important to show off our winning of an ERC starters grant...
November 29, 2013
We got 1.63 million Euros for our project: Beads on String Genomics: Experimental Toolbox for Unmasking Genetic / Epigenetic Variation in Genomic DNA and Chromatin.

Optical detection of epigenetic marks: sensitive quantification and direct imaging of individual hydroxymethylcytosine bases
July 15, 2013
The first 100% independent work from our lab and the first of its kind observation of individual epigenetic modifications in genomic DNA. Just published on the front cover of Chemical Communications.

Beyond sequencing: optical mapping of DNA in the age of nanotechnology and nanoscopy
July 15, 2013
We were very happy to be invited to write this short review for a special issue on bio-nano-technology in Current opinion in biotechnology. It really gave us the opportunity to see what was going on in the field and to refine our opinion on where this technology is going.


.jpeg)







































